Iris flower dataset¶
The iris flower dataset is a common dataset used in machine learning.
It has been created Ronald Fisher in 1936. It contains the petal length, petal width, sepal length and sepal width of 150 iris flowers from 3 different species.
Dataset has been downloaded from Kaggle.
To go through this example, you need to install AutoClassWrapper:
$ python3 -m pip install autoclasswrapper
AutoClass C also needs to be installed locally and available in path.
Here is a quick solution for a Linux Bash shell:
wget https://ti.arc.nasa.gov/m/project/autoclass/autoclass-c-3-3-6.tar.gz
tar zxvf autoclass-c-3-3-6.tar.gz
rm -f autoclass-c-3-3-6.tar.gz
export PATH=$PATH:$(pwd)/autoclass-c
# if you use a 64-bit operating system,
# you also need to install the standard 32-bit C libraries:
# sudo apt-get install -y libc6-i386
[1]:
from pathlib import Path
import sys
import time
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
import numpy as np
import pandas as pd
%matplotlib inline
print("Python:", sys.version)
print("matplotlib:", matplotlib.__version__)
print("numpy:", np.__version__)
print("pandas:", pd.__version__)
import autoclasswrapper as wrapper
print("AutoClassWrapper:", wrapper.__version__)
version = sys.version_info
if not ((version.major >= 3) and (version.minor >= 6)):
sys.exit("Need Python>=3.6")
Python: 3.7.1 | packaged by conda-forge | (default, Feb 26 2019, 04:48:14)
[GCC 7.3.0]
matplotlib: 3.0.3
numpy: 1.16.2
pandas: 0.24.1
AutoClassWrapper: 1.4.1
Dataset preparation¶
[2]:
df = pd.read_csv("iris.csv", index_col="Id")
df.head()
[2]:
| SepalLengthCm | SepalWidthCm | PetalLengthCm | PetalWidthCm | Species | |
|---|---|---|---|---|---|
| Id | |||||
| 1 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 3 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 5 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
[3]:
df.describe(include='all').T
[3]:
| count | unique | top | freq | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SepalLengthCm | 150 | NaN | NaN | NaN | 5.84333 | 0.828066 | 4.3 | 5.1 | 5.8 | 6.4 | 7.9 |
| SepalWidthCm | 150 | NaN | NaN | NaN | 3.054 | 0.433594 | 2 | 2.8 | 3 | 3.3 | 4.4 |
| PetalLengthCm | 150 | NaN | NaN | NaN | 3.75867 | 1.76442 | 1 | 1.6 | 4.35 | 5.1 | 6.9 |
| PetalWidthCm | 150 | NaN | NaN | NaN | 1.19867 | 0.763161 | 0.1 | 0.3 | 1.3 | 1.8 | 2.5 |
| Species | 150 | 3 | Iris-versicolor | 50 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
Add discrete values¶
Apart iris species, data in this dataset are numerical values only.
To demonstrate the ability of AutoClass C to handle discrete values, we will convert PetalWidthCm column to discrete categorical values.
[4]:
def categorize(value):
if value <= 0.75:
return "small"
elif 0.75 < value <= 1.75:
return "medium"
elif 1.75 < value:
return "large"
[5]:
df["PetalWidthCat"] = df["PetalWidthCm"].apply(categorize)
df.head()
[5]:
| SepalLengthCm | SepalWidthCm | PetalLengthCm | PetalWidthCm | Species | PetalWidthCat | |
|---|---|---|---|---|---|---|
| Id | ||||||
| 1 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa | small |
| 2 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa | small |
| 3 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa | small |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa | small |
| 5 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa | small |
Add missing values¶
To demonstrate the ability of AutoClass C to handle missing values, we will delete some values.
[6]:
df.loc[1, "SepalLengthCm"] = np.nan
df.loc[2, "SepalWidthCm"] = np.nan
df.loc[3, "PetalLengthCm"] = np.nan
df.head()
[6]:
| SepalLengthCm | SepalWidthCm | PetalLengthCm | PetalWidthCm | Species | PetalWidthCat | |
|---|---|---|---|---|---|---|
| Id | ||||||
| 1 | NaN | 3.5 | 1.4 | 0.2 | Iris-setosa | small |
| 2 | 4.9 | NaN | 1.4 | 0.2 | Iris-setosa | small |
| 3 | 4.7 | 3.2 | NaN | 0.2 | Iris-setosa | small |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa | small |
| 5 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa | small |
Save dataset in two different files. One with real values and the other one with discrete values (column PetalWidthCat). Missing values must encoded with nothing.
[7]:
df.drop(["Species", "PetalWidthCm", "PetalWidthCat"], axis=1).to_csv("iris_real.tsv", sep="\t", header=True)
!head iris_real.tsv
Id SepalLengthCm SepalWidthCm PetalLengthCm
1 3.5 1.4
2 4.9 1.4
3 4.7 3.2
4 4.6 3.1 1.5
5 5.0 3.6 1.4
6 5.4 3.9 1.7
7 4.6 3.4 1.4
8 5.0 3.4 1.5
9 4.4 2.9 1.4
[8]:
df["PetalWidthCat"].to_csv("iris_discrete.tsv", sep="\t", header=True)
!head iris_discrete.tsv
Id PetalWidthCat
1 small
2 small
3 small
4 small
5 small
6 small
7 small
8 small
9 small
Step 1 - prepare input files¶
[9]:
# Create object to prepare dataset.
clust = wrapper.Input()
# Load datasets from tsv files.
clust.add_input_data("iris_real.tsv", "real scalar")
clust.add_input_data("iris_discrete.tsv", "discrete")
# Prepare input data:
# - create a final dataframe
# - merge datasets if multiple inputs
clust.prepare_input_data()
# Create files needed by AutoClass.
clust.create_db2_file()
clust.create_hd2_file()
clust.create_model_file()
# We wanted reproducible results to ease documentation.
# But bear in mind, that this parameter is not advised by authors of AutoClass C in production run.
# Use clust.create_sparams_file() instead.
clust.create_sparams_file(reproducible_run=True)
clust.create_rparams_file()
2019-07-07 19:07:58 INFO Reading data file 'iris_real.tsv' as 'real scalar' with error 0.01
2019-07-07 19:07:58 INFO Detected encoding: ascii
2019-07-07 19:07:59 INFO Found 150 rows and 4 columns
2019-07-07 19:07:59 DEBUG Checking column names
2019-07-07 19:07:59 DEBUG Index name 'Id'
2019-07-07 19:07:59 DEBUG Column name 'SepalLengthCm'
2019-07-07 19:07:59 DEBUG Column name 'SepalWidthCm'
2019-07-07 19:07:59 DEBUG Column name 'PetalLengthCm'
2019-07-07 19:07:59 INFO Checking data format
2019-07-07 19:07:59 INFO Column 'SepalLengthCm'
2019-07-07 19:07:59 INFO count 149.000000
2019-07-07 19:07:59 INFO mean 5.848322
2019-07-07 19:07:59 INFO std 0.828594
2019-07-07 19:07:59 INFO min 4.300000
2019-07-07 19:07:59 INFO 50% 5.800000
2019-07-07 19:07:59 INFO max 7.900000
2019-07-07 19:07:59 INFO ---
2019-07-07 19:07:59 INFO Column 'SepalWidthCm'
2019-07-07 19:07:59 INFO count 149.000000
2019-07-07 19:07:59 INFO mean 3.054362
2019-07-07 19:07:59 INFO std 0.435034
2019-07-07 19:07:59 INFO min 2.000000
2019-07-07 19:07:59 INFO 50% 3.000000
2019-07-07 19:07:59 INFO max 4.400000
2019-07-07 19:07:59 INFO ---
2019-07-07 19:07:59 INFO Column 'PetalLengthCm'
2019-07-07 19:07:59 INFO count 149.000000
2019-07-07 19:07:59 INFO mean 3.775168
2019-07-07 19:07:59 INFO std 1.758720
2019-07-07 19:07:59 INFO min 1.000000
2019-07-07 19:07:59 INFO 50% 4.400000
2019-07-07 19:07:59 INFO max 6.900000
2019-07-07 19:07:59 INFO ---
2019-07-07 19:07:59 INFO Reading data file 'iris_discrete.tsv' as 'discrete'
2019-07-07 19:07:59 INFO Detected encoding: ascii
2019-07-07 19:07:59 INFO Found 150 rows and 2 columns
2019-07-07 19:07:59 DEBUG Checking column names
2019-07-07 19:07:59 DEBUG Index name 'Id'
2019-07-07 19:07:59 DEBUG Column name 'PetalWidthCat'
2019-07-07 19:07:59 INFO Checking data format
2019-07-07 19:07:59 INFO Column 'PetalWidthCat': 3 different values
2019-07-07 19:07:59 INFO Preparing input data
2019-07-07 19:07:59 INFO Final dataframe has 150 lines and 5 columns
2019-07-07 19:07:59 INFO Searching for missing values
2019-07-07 19:07:59 WARNING Missing values found in column: SepalLengthCm
2019-07-07 19:07:59 WARNING Missing values found in column: SepalWidthCm
2019-07-07 19:07:59 WARNING Missing values found in column: PetalLengthCm
2019-07-07 19:07:59 INFO Writing autoclass.db2 file
2019-07-07 19:07:59 INFO If any, missing values will be encoded as '?'
2019-07-07 19:07:59 DEBUG Writing autoclass.tsv file [for later use]
2019-07-07 19:07:59 INFO Writing .hd2 file
2019-07-07 19:07:59 INFO Writing .model file
2019-07-07 19:07:59 INFO Writing .s-params file
2019-07-07 19:07:59 INFO Writing .r-params file
Step 2 - prepare run script & run autoclass¶
[10]:
# Clean previous status file and results if a classification has already been performed.
!rm -f autoclass-run-* *.results-bin
# Search autoclass in path.
wrapper.search_autoclass_in_path()
# Create object to run AutoClass.
run = wrapper.Run()
# Prepare run script.
run.create_run_file()
# Run AutoClass.
run.run()
2019-07-07 19:08:02 INFO AutoClass C executable found in /home/pierre/.soft/bin/autoclass
2019-07-07 19:08:02 INFO Writing run file
2019-07-07 19:08:02 INFO AutoClass C executable found in /home/pierre/.soft/bin/autoclass
2019-07-07 19:08:02 INFO AutoClass C version: AUTOCLASS C (version 3.3.6unx)
2019-07-07 19:08:02 INFO Running clustering...
Step 3 - parse and format results¶
[11]:
timer = 0
step = 2
while not Path("autoclass-run-success").exists():
timer += step
sys.stdout.write("\r")
sys.stdout.write(f"Time: {timer} sec.")
sys.stdout.flush()
time.sleep(step)
results = wrapper.Output()
results.extract_results()
results.aggregate_input_data()
results.write_cdt()
results.write_cdt(with_proba=True)
results.write_class_stats()
results.write_dendrogram()
2019-07-07 19:08:05 INFO Extracting autoclass results
2019-07-07 19:08:05 INFO Found 150 cases classified in 4 classes
2019-07-07 19:08:05 INFO Aggregating input data
2019-07-07 19:08:05 INFO Writing classes + probabilities .tsv file
2019-07-07 19:08:05 INFO Writing .cdt file
2019-07-07 19:08:05 INFO Writing .cdt file (with probabilities)
2019-07-07 19:08:05 INFO Writing class statistics
2019-07-07 19:08:05 INFO Writing dendrogram
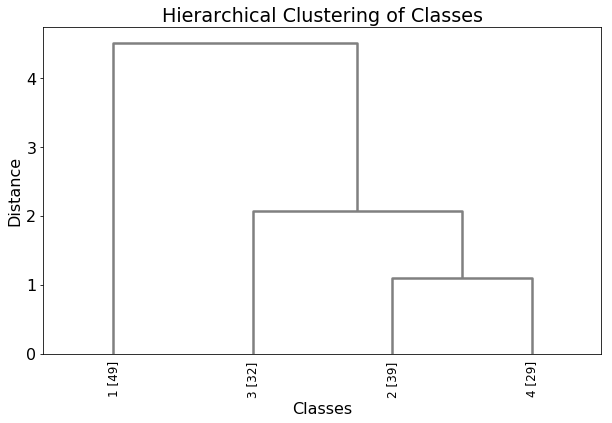
For comparison, add class number to original dataset.
[12]:
df_class = pd.read_csv("autoclass_out.tsv", sep="\t", index_col="Id")
df = pd.concat([df, df_class["main-class"]], axis=1, join="outer")
df.head()
[12]:
| SepalLengthCm | SepalWidthCm | PetalLengthCm | PetalWidthCm | Species | PetalWidthCat | main-class | |
|---|---|---|---|---|---|---|---|
| Id | |||||||
| 1 | NaN | 3.5 | 1.4 | 0.2 | Iris-setosa | small | 1 |
| 2 | 4.9 | NaN | 1.4 | 0.2 | Iris-setosa | small | 1 |
| 3 | 4.7 | 3.2 | NaN | 0.2 | Iris-setosa | small | 1 |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa | small | 1 |
| 5 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa | small | 1 |
Compute class distribution for iris species¶
[13]:
pd.pivot_table(df, index=["Species"], columns=["main-class"], values=[], aggfunc=len, fill_value=0)
[13]:
| main-class | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Species | ||||
| Iris-setosa | 50 | 0 | 0 | 0 |
| Iris-versicolor | 0 | 23 | 0 | 27 |
| Iris-virginica | 0 | 16 | 32 | 2 |
The setosa species is found only in cluster 1. Note that missing values did not interfere in the classification of the 3 first flowers as setosa.
The versicolor species is found in cluster 2 and 4.
The virginica species is found mainly in cluster 3 but also in cluster 2 and 4.
AutoClass-C determines automatically what is the optimal number of classes. It’s always a good idea to analyse the final results to check if some cluster can be merged (for instance cluster 2 and 4).